CIS 1.5 (Science Section)
Professor Langsam
Assignment #6
The Genetic Code[1]
Deoxyribonucleic
acid, or DNA, is a
molecule that contains the instructions used in the development and functioning
of all known living organisms. The main role of DNA is the long-term storage of
information and it is often compared to a set of blueprints, since DNA contains
the instructions needed to construct other components of cells, such as proteins
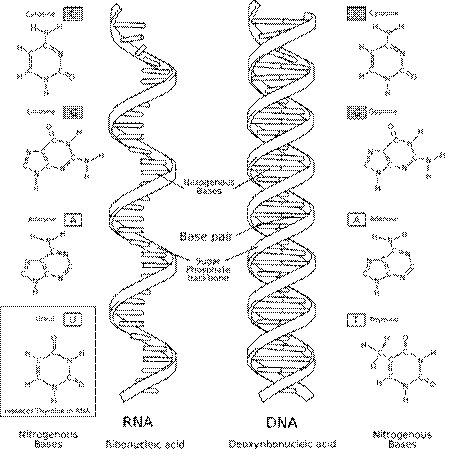
and RNA molecules. DNA is a molecule in the form of a double helix
(twisted-ladder) Attached to the backbone are of four types of molecules called
bases – these form the rungs of the ladder. It is the sequence of these four
bases along the backbone that encodes information. The four bases found in DNA
are adenine (abbreviated A), cytosine (C), guanine (G) and thymine (T). RNA
is similar with the base uracil (U) rather than thymine. See the figure below.
The genome of an
organism is inscribed in the DNA. The portion of the genome that codes for a
protein or an RNA molecule is referred to as a gene. The genetic code is
the set of rules by which information encoded in genetic material (DNA or RNA
sequences) is translated into proteins (amino acid sequences) by living cells.
Specifically, the code defines a mapping between tri-nucleotide sequences
called codons and amino acids; every triplet of nucleotides in a nucleic
acid sequence specifies a single amino acid
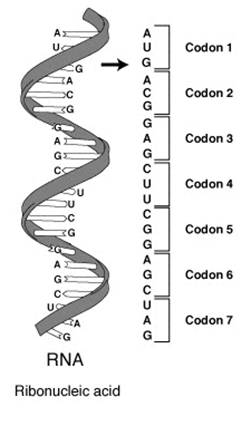
Since there are 4 possible bases (A, C, G, T or U) and each
codon consists of 3 bases there are
4³ = 64 different combinations possible with a triplet codon of three
nucleotides. If, for example, an RNA sequence, UUUAAACCC is considered, there
are three codons, namely, UUU, AAA and CCC, each of which specifies one amino
acid. This RNA sequence will be translated into an amino acid sequence, three
amino acids long.
The
standard genetic code is shown in the following tables. Table 1 shows what
amino acid each of the 64 codons specifies. Table 2 shows what codons specify
each of the 20 standard amino acids involved in translation. These are called
forward and reverse codon tables, respectively. For example, the codon AAU
represents the amino acid asparagine, and UGU and UGC represent cysteine
(standard three-letter designations, Asn and Cys respectively). Note several
codons can code for the same amino acid.
Table 1: RNA Codon table - This table shows the 64 codons and the amino acid each codon codes for. |
|
|
2nd base |
||||
|
U |
C |
A |
G |
||
|
1st |
U |
UUU (Phe/F)Phenylalanine |
UCU (Ser/S)Serine |
UAU (Tyr/Y)Tyrosine |
UGU (Cys/C)Cysteine |
|
C |
CUU (Leu/L)Leucine |
CCU (Pro/P)Proline |
CAU (His/H)Histidine |
CGU (Arg/R)Arginine |
|
|
A |
AUU (Ile/I)Isoleucine |
ACU (Thr/T)Threonine |
AAU (Asn/N)Asparagine |
AGU (Ser/S)Serine |
|
|
G |
GUU (Val/V)Valine |
GCU ( |
GAU (Asp/D)Aspartic acid |
GGU (Gly/G)Glycine |
|
Table 2: Inverse table |
|
|
GCU, GCC, GCA, GCG |
Leu/L |
UUA, UUG, CUU, CUC, CUA,
CUG |
|
Arg/R |
CGU, CGC, CGA, CGG, AGA,
AGG |
Lys/K |
AAA, AAG |
|
Asn/N |
AAU, AAC |
Met/M |
AUG |
|
Asp/D |
GAU, GAC |
Phe/F |
UUU, UUC |
|
Cys/C |
UGU, UGC |
Pro/P |
CCU, CCC, CCA, CCG |
|
Gln/Q |
CAA, CAG |
Ser/S |
UCU, UCC, UCA, UCG, AGU,
AGC |
|
Glu/E |
GAA, GAG |
Thr/T |
ACU, ACC, ACA, ACG |
|
Gly/G |
GGU, GGC, GGA, GGG |
Trp/W |
UGG |
|
His/H |
CAU, CAC |
Tyr/Y |
UAU, UAC |
|
Ile/I |
AUU, AUC, AUA |
Val/V |
GUU, GUC, GUA, GUG |
|
START |
AUG |
STOP |
UAG, UGA, UAA |
Note that a codon
is defined by the initial nucleotide from which translation starts. For
example, the string GGGAAACCC, if read from the first position, contains the
codons GGG, AAA and CCC; and if read from the second position, it contains the
codons GGA and AAC; if read starting from the third position, GAA and ACC.
Partial codons have been ignored in this example. Every sequence can thus be
read in three reading frames, each of which will produce a different
amino acid sequence (in the given example, Gly-Lys-Pro, Gly-Asp, or Glu-Thr,
respectively).
The actual frame of a protein is translated in is defined by a start codon, usually the first AUG codon in the mRNA sequence. There are three stop codons (UAG, UGA, UAA) which signal then end of a sequence.
Proteins are large organic compounds made of amino acids arranged in a linear chain. The sequence of amino acids in a protein is defined by a gene and encoded in the genetic code. Proteins are essential parts of organisms and participate in every process within cells.
The entire process may be represented by the following diagram:
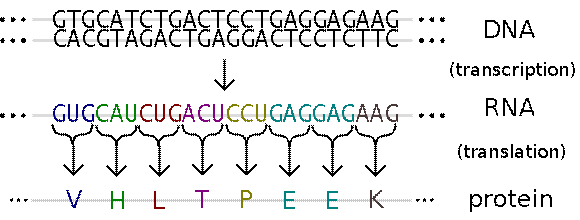
Write a program that examines a sequence of bases in a strand of RNA and prints the amino acid sequence for each protein coded within that strand. For example, given the following portion of an RNA strand:
…AAUUGUAUGAAAUUUCCUGAAUAUUAGGAUGCUCAAAAAAUGUGGUUUUUGUUGGAACAAGACUAAUACUUUU…
Your program should print the following:
Protein #1: KFPEY
Protein #2: WFLLEQD
Note that each protein begins with a start codon and ends with a stop codon. The sequence in between the stop codon and the next start codon does not code for any protein and is known as junk DNA. An actual protein may be hundreds or thousands of amino acids long. Also note that we are using the one letter abbreviations of the 20 amino acids that make up all proteins.
Strategy
- Create a data file that contains the information given in Table 2. The table should have one entry per line:
GCU A
GCC A
GCA A
GCG A
CGU R
CGC R
.
. .
- Write a function, readRNACodonTable, that reads this data file into a two dimensional array.
- Write a function, sort, that sorts this array in codon order.
- Write a function, codonLookup, that given a three character codon returns the one-letter abbreviation for the corresponding amino acid by searching the table you created in step 2.
- The main function should:
- Read the RNA strand from a file one character at a time until a start codon is encountered.
- Continue to read the RNA strand three characters at a time and call the function codonLookup to identify the corresponding amino acid.
- Print the one-letter abbreviation returned by codonLookup.
- Repeat step 5b-c until a stop codon is returned.
- Continue scanning the RNA strand (steps 5a-d) until the end of the strand is reached.
Extra Credit:
- Have the main function print the junk DNA sequence.
- Write a reverse function that accepts an amino acid sequence and prints each of the possible codon sequences that would code this protein. (There may be more than one. Why?) – Use this function on each protein that you have found in the original assignment.
Data: (copy this carefully – you may also download it from the web at:
http://eilat.sci.brooklyn.cuny.edu/cis1_5/CISClassPage.htm )
…AACAAUAUUAUGCAACAGUGUCCUCCCUUAUGAGCGUGUGGUUAGAAUUGUAUGAAAUUUCCUGAAUAUUAGGAUGCUCAAAAAAUGUGGUUUUUGUUGGAACAAGACUAAUACUUUUUUGUUGAUGAGAAUGAAACCCCCCAAAUUUAGAGCUGCCAGACAUCAACCCUUUAAACCCCCCUAGUUUCCCAAA…